library(tidyverse)
library(magrittr)
link_list_dk <- list(
"1996_atc_code_data.txt" = "https://medstat.dk/da/download/file/MTk5Nl9hdGNfY29kZV9kYXRhLnR4dA==",
"1997_atc_code_data.txt" = "https://medstat.dk/da/download/file/MTk5N19hdGNfY29kZV9kYXRhLnR4dA==",
"1998_atc_code_data.txt" = "https://medstat.dk/da/download/file/MTk5OF9hdGNfY29kZV9kYXRhLnR4dA==",
"1999_atc_code_data.txt" = "https://medstat.dk/da/download/file/MTk5OV9hdGNfY29kZV9kYXRhLnR4dA==",
"2000_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwMF9hdGNfY29kZV9kYXRhLnR4dA==",
"2001_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwMV9hdGNfY29kZV9kYXRhLnR4dA==",
"2002_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwMl9hdGNfY29kZV9kYXRhLnR4dA==",
"2003_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwM19hdGNfY29kZV9kYXRhLnR4dA==",
"2004_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwNF9hdGNfY29kZV9kYXRhLnR4dA==",
"2006_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwNl9hdGNfY29kZV9kYXRhLnR4dA==",
"2007_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwN19hdGNfY29kZV9kYXRhLnR4dA==",
"2008_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwOF9hdGNfY29kZV9kYXRhLnR4dA==",
"2009_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwOV9hdGNfY29kZV9kYXRhLnR4dA==",
"2010_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxMF9hdGNfY29kZV9kYXRhLnR4dA==",
"2011_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxMV9hdGNfY29kZV9kYXRhLnR4dA==",
"2012_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxMl9hdGNfY29kZV9kYXRhLnR4dA==",
"2013_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxM19hdGNfY29kZV9kYXRhLnR4dA==",
"2014_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxNF9hdGNfY29kZV9kYXRhLnR4dA==",
"2015_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxNV9hdGNfY29kZV9kYXRhLnR4dA==",
"2016_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxNl9hdGNfY29kZV9kYXRhLnR4dA==",
"2017_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxN19hdGNfY29kZV9kYXRhLnR4dA==",
"2018_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxOF9hdGNfY29kZV9kYXRhLnR4dA==",
"2019_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxOV9hdGNfY29kZV9kYXRhLnR4dA==",
"2020_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAyMF9hdGNfY29kZV9kYXRhLnR4dA==",
"2021_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAyMV9hdGNfY29kZV9kYXRhLnR4dA==",
"2022_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAyMl9hdGNfY29kZV9kYXRhLnR4dA==",
"atc_code_text.txt" = "https://medstat.dk/da/download/file/YXRjX2NvZGVfdGV4dC50eHQ=",
"atc_groups.txt" = "https://medstat.dk/da/download/file/YXRjX2dyb3Vwcy50eHQ=",
"population_data.txt" = "https://medstat.dk/da/download/file/cG9wdWxhdGlvbl9kYXRhLnR4dA=="
)
# ATC codes are stored in files
seq_atc <- c(1:26)
atc_code_data_list <- link_list_dk[seq_atc]
# drugs names
eng_drug <- read_delim(link_list_dk[["atc_groups.txt"]], delim = ";", col_names = c(paste0("V", c(1:6))), col_types = cols(V1 = col_character(), V2 = col_character(), V3 = col_character(), V4 = col_character(), V5 = col_character(), V6 = col_character())) %>%
# keep drug classes labels in English
filter(V5 == "1") %>%
select(ATC = V1,
drug = V2,
unit_dk = V4)
# drugs data
atc_data <- map(atc_code_data_list, ~read_delim(file = .x, delim = ";", trim_ws = T, col_names = c(paste0("V", c(1:14))), col_types = cols(V1 = col_character(), V2 = col_character(), V3 = col_character(), V4 = col_character(), V5 = col_character(),V6 = col_character(), V7 = col_character(), V8 = col_character(), V9 = col_character(), V10 = col_character(), V11 = col_character(), V12 = col_character(), V13 = col_character(), V14 = col_character()))) %>%
# bind atc_data from all years
bind_rows()
# population data
pop_data <- read_delim(link_list_dk[["population_data.txt"]], delim = ";", col_names = c(paste0("V", c(1:7))), col_types = cols(V1 = col_character(), V2 = col_character(), V3 = col_character(), V4 = col_character(), V5 = col_character(), V6 = col_character(), V7 = col_character())) %>%
# rename and keep columns
select(year = V1,
region_text = V2,
region = V3,
gender = V4,
age = V5,
denominator_per_year = V6) %>%
# human-reading friendly label on sex categories
mutate(
gender_text = case_when(
gender == "1" ~ "men",
gender == "2" ~ "women",
T ~ as.character(gender)
)
) %>%
# make numeric variables
mutate_at(vars(year, age, denominator_per_year), as.numeric) %>%
arrange(year, age, region, gender)
atc_data %<>%
# rename and keep columns
rename(
ATC = V1,
year = V2,
sector = V3,
region = V4,
gender = V5, # 0 - both; 1 - male; 2 - female; A - age in categories
age = V6,
number_of_people = V7,
patients_per_1000_inhabitants = V8,
turnover = V9,
regional_grant_paid = V10,
quantity_sold_1000_units = V11,
quantity_sold_units_per_unit_1000_inhabitants_per_day = V12,
percentage_of_sales_in_the_primary_sector = V13
)
atc_data %<>%
# clean columns names and set-up labels
mutate(
year = as.numeric(year),
region_text = case_when(
region == "0" ~ "DK",
region == "1" ~ "Region Hovedstaden",
region == "2" ~ "Region Midtjylland",
region == "3" ~ "Region Nordjylland",
region == "4" ~ "Region Sjælland",
region == "5" ~ "Region Syddanmark",
T ~ NA_character_
),
gender_text = case_when(
gender == "0" ~ "both sexes",
gender == "1" ~ "men",
gender == "2" ~ "women",
T ~ as.character(gender)
)
) %>%
mutate_at(vars(turnover, regional_grant_paid, quantity_sold_1000_units, quantity_sold_units_per_unit_1000_inhabitants_per_day, number_of_people, patients_per_1000_inhabitants), as.numeric) %>%
select(-V14) %>%
filter(
region == "0",
sector == "0",
gender == "2"
)
atc_data %<>%
# deal with non-numeric age in groups
filter(
str_detect(age, "[0-9][0-9][0-9]")
) %>%
mutate(
age = parse_number(age)
) %>%
select(year, ATC, gender, age, number_of_people, patients_per_1000_inhabitants, region_text, gender_text)
regex_pregabalin <- "^N02BF02$"
atc_data %<>% filter(str_detect(ATC, regex_pregabalin))
atc_data %<>% left_join(eng_drug)
atc_data %<>% left_join(pop_data)
# group to the same categories as in Swedish data
atc_data_se <- readxl::read_xlsx(path = "C:/Users/au595736/OneDrive - Aarhus Universitet/ISPOR 2023/Statistikdatabasen_2023-03-15 10_54_00.xlsx", sheet = 1) %>%
pivot_longer(cols = c("2006":"2022")) %>%
mutate(
name = as.numeric(name),
country = "Sweden"
)
atc_data %<>% mutate(
age_cat = cut(age, c(-Inf, 14, 19, 24, 29, 34, 39, 44, 49, Inf))
) %>%
group_by(age_cat, year) %>%
mutate(
number_of_people_age_cat = sum(number_of_people),
denominator_per_year_age_cat = sum(denominator_per_year),
patients_per_1000_inhabitants_age_cat = (number_of_people_age_cat/denominator_per_year_age_cat)*1000
) %>%
ungroup() %>%
filter(
age_cat != "(-Inf,14]",
age_cat != "(49, Inf]"
) %>%
mutate(
age_cat = case_when(
age_cat == "(14,19]" ~ "15-19",
age_cat == "(19,24]" ~ "20-24",
age_cat == "(24,29]" ~ "25-29",
age_cat == "(29,34]" ~ "30-34",
age_cat == "(34,39]" ~ "35-39",
age_cat == "(39,44]" ~ "40-44",
age_cat == "(44,49]" ~ "45-49"
),
country = "Denmark"
) %>%
distinct(age_cat, year, number_of_people_age_cat, denominator_per_year_age_cat, patients_per_1000_inhabitants_age_cat)
atc <- full_join(atc_data, atc_data_se, by = c("year" = "name", "age_cat" = "Ålder", "patients_per_1000_inhabitants_age_cat" = "value", "country" = "country")) %>%
select(year, age_cat, patients_per_1000_inhabitants_age_cat, country)
ggplot <- atc %>%
# filter(!is.na(patients_per_1000_inhabitants_age_cat)) %>%
mutate(label = if_else(year == 2022, as.character(age_cat), NA_character_),
value = case_when(
age_cat == "30-34" ~"#3E9286",
age_cat == "35-39" ~ "#E8B88A",
T ~ "gray"
)) %>%
ggplot(aes(x = year, y = patients_per_1000_inhabitants_age_cat, group = age_cat, color = value)) +
geom_line() +
facet_grid(cols = vars(country), scales = "free", drop = F) +
theme_light(base_size = 14, base_family = "Ubuntu") +
scale_x_continuous(breaks = c(seq(2004, 2022, 5))) +
scale_color_identity() +
theme(plot.caption = element_text(hjust = 0, size = 10),
legend.position = "none",
panel.spacing = unit(0.8, "cm")) +
labs(y = "Female patients of reproductive age\nper 1,000 women in the population", title = "Pregabalin utilization in women", subtitle = "aged between 15 and 49 years in DK and SE\n2004-2022", caption = "Elena Dudukina | @evpatora") +
ggrepel::geom_label_repel(aes(label = label), na.rm = TRUE, nudge_x = 3, hjust = 0.5, direction = "y", segment.size = 0.1, segment.colour = "black", show.legend = F, family = "Ubuntu")
ggsave(filename = paste0(Sys.Date(), "_pregabalin use in DK and SE.pdf"), plot = ggplot, path = "C:/Users/au595736/OneDrive - Aarhus Universitet/ISPOR 2023/", dpi = 900, height = 6, width = 8, device = cairo_pdf)Prenatal Exposure to Antiseizure Medication and Risk of Neurodevelopmental Outcomes: evidence from the Nordics
ISPOR-AU General Meeting, March 2023
Prenatal exposure to antiseizure medication
What is known?
SCAN-AED
Nordics: Denmark, Finland, Iceland, Norway, and Sweden:
4 494 926 participants
Prenatal exposure to antiseizure medication: topiramate, valproate, lamotrigine, carbamazepine, valproate, pregabalin, gabapentin, mono- and duotherapies
Comparators:
Children born from antiseizure therapy-unexposed pregnancies of women with epilepsy
Children born antiseizure therapy-unexposed of women in the total population
Outcomes:
Autism spectrum disorder
Intellectual disability
Any neurodevelopmental disorder
Previous results from the Nordics
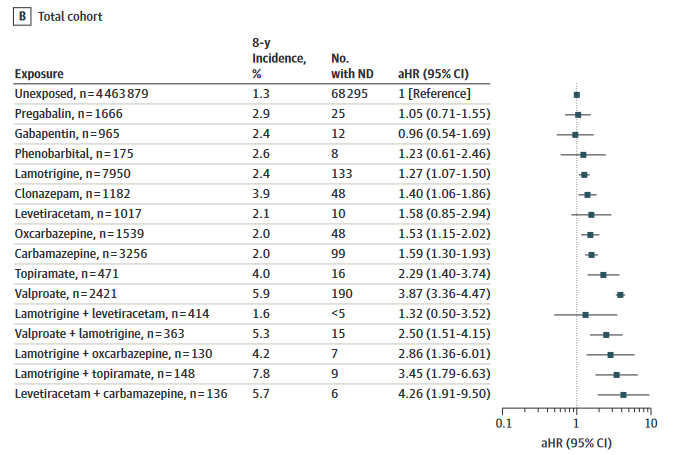
No increased risks were identified for levetiracetam, gabapentin, pregabalin, or phenobarbital
Topiramate and valproate in monotherapy were associated with a 2- to 4-fold increased risk of autism spectrum disorders and intellectual disability1
Further questions
Use of active comparators for most used antiepileptic medication: pregabalin
Exposure unrestricted to mono-therapy (any prenatal exposure to pregabalin)
 vs
vs 
Pregabalin indications
Approved indications for pregabalin (in the European Union):
Epilepsy
Neuropathic pain
Generalized anxiety disorder (GAD)

Previous evidence on prenatal pregabalin exposure and adverse outcomes
- European Network of Teratology Information Services (ENTIS):
- 3-fold increased risk of any major non-chromosomal congenital malformation
- Medicaid beneficiaries + MarketScan in the US:
- Pooled RR=1.33 (95% CI: 0.83-2.15); interpreted as no association
- Confounding by indication is a major concern since most of the earlier studies compared exposure to pregabalin with no exposure to AED
- Postnatal outcomes are rarely investigated
- Particular postnatal outcomes are of interest, eg, ADHD

Country-level data on pregabalin use in women
Pregabalin is an antiepileptic drug (AED), prescribed to approximately 0.5 per thousand pregnant women in Europe, with increasing use after 2010.
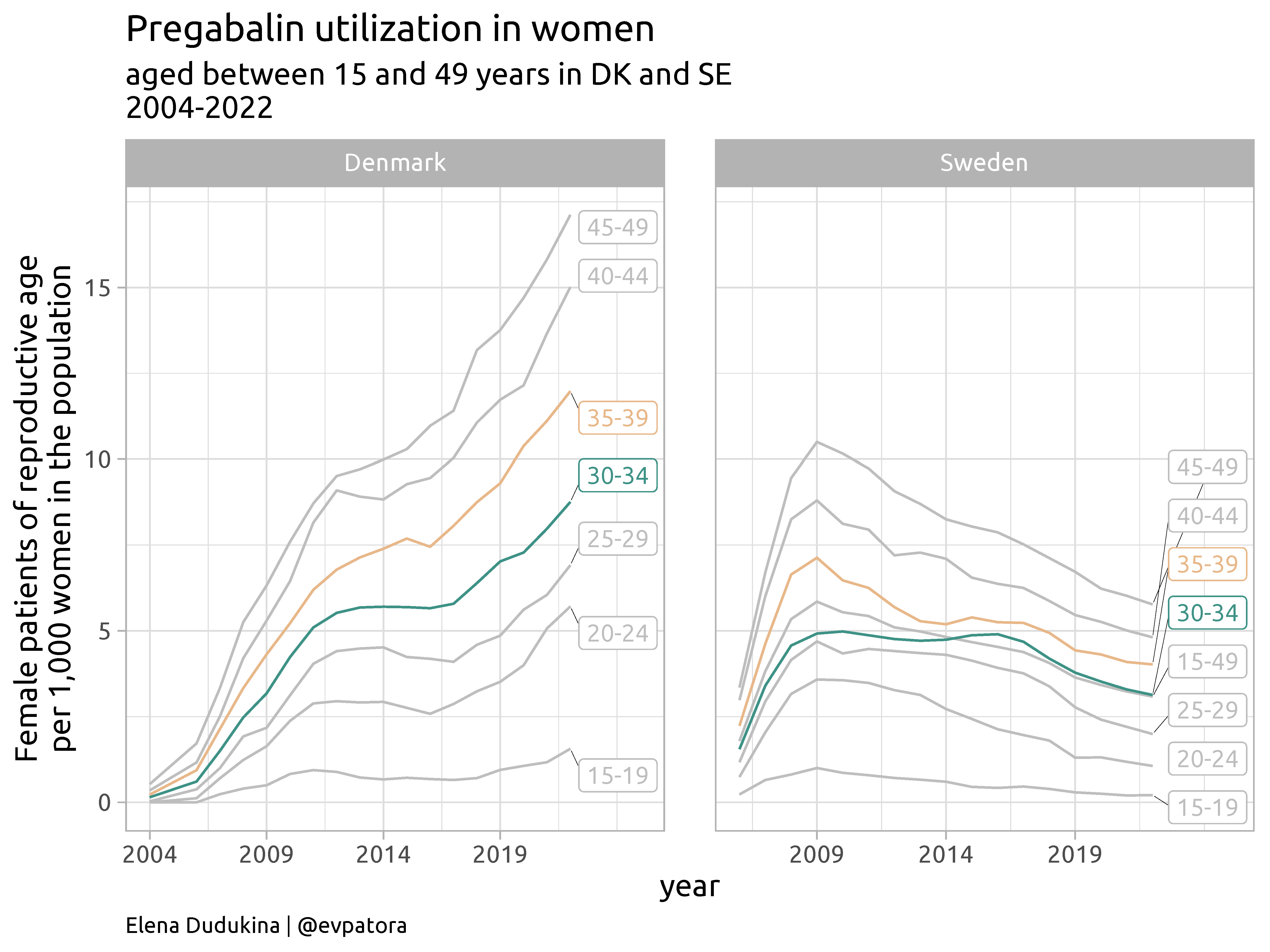
Pregabalin and neurodevelopment study in the Nordics
Setting
- Non-interventional study was a post-authorisation safety (PAS) study conducted as a commitment to the European Medicines Agency (EUPAS27339)
- Participants:
- Denmark
- Finland
- Sweden
- Norway
- Time period: 2005-2016
- Population-based
- All births identified in the respective medical birth registers from 01 January 2005 through 31 December 2015


Pregabalin and neurodevelopment study in the Nordics
Exposure
- Any-trimester pregabalin exposure as at least one dispensing of pregabalin from LMP-90 days to the day before date of birth
- Pregabalin monotherapy in sensitivity analysis

Comparators
- No exposure to pregagbalin or other antiepileptics or active comparators
- Active comparators
- Lamotrigine
![]()
- Duloxetine
![]()
- Lamotrigine and/or duloxetine
![]() and/or
and/or ![]()
- Lamotrigine
Pregabalin and neurodevelopment study in the Nordics
Design
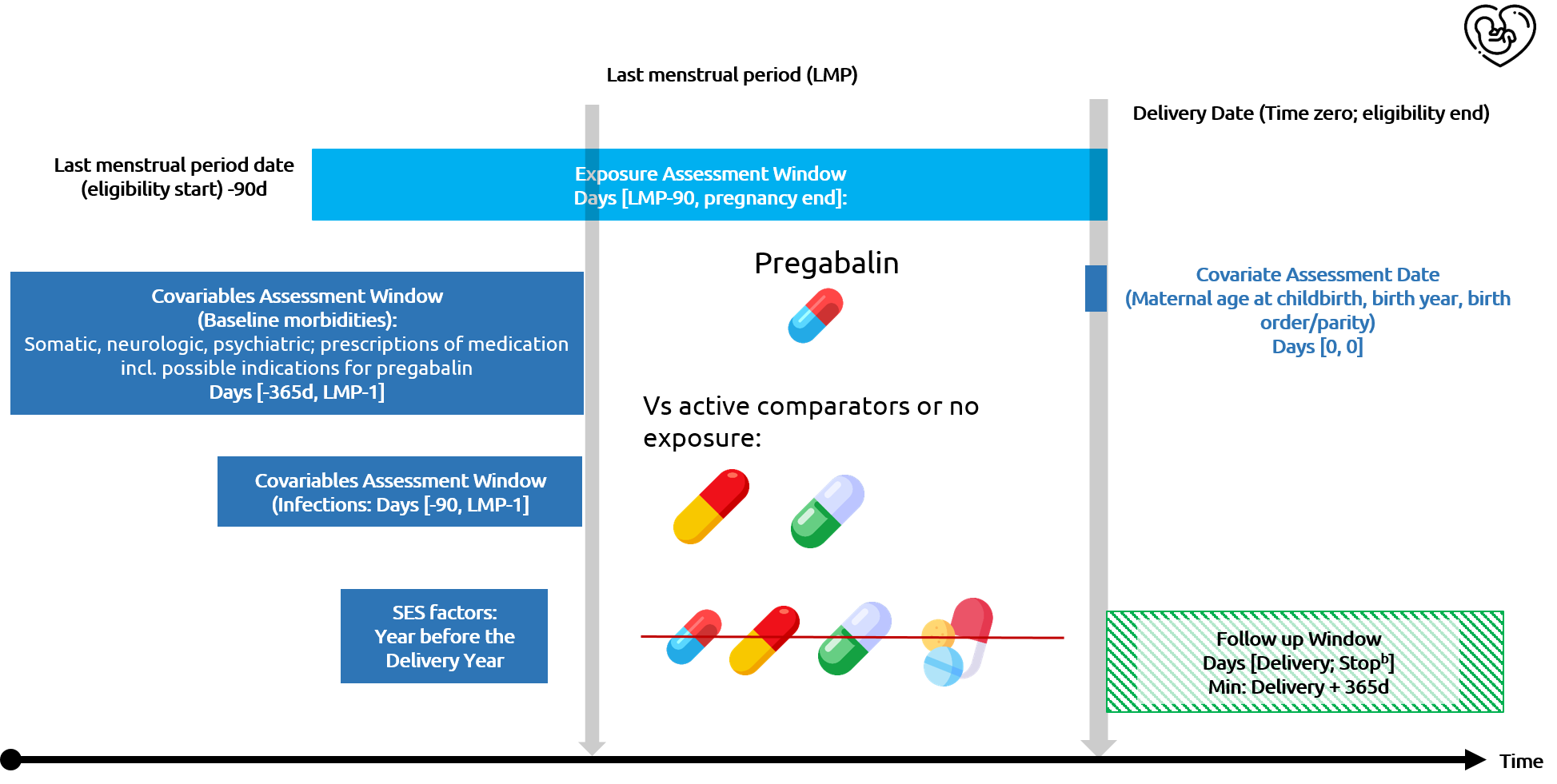
Pregabalin and neurodevelopment study in the Nordics
Covariables and confounding control




Pregabalin and neurodevelopment study in the Nordics
Covariables
- All covariables were available for at least 12 months before the end of the earliest identified pregnancy
- Maternal pre-pregnancy comorbidities using diagnostic codes from the national patient registries
- Maternal pre-pregnancy medication use from the national prescription registers



Pregabalin and neurodevelopment study in the Nordics
Descriptive variables:
- Caesarean delivery
![]()
- Children’s sex distributions
![]()
- Distributions of potential indications for pregabalin use inferred by recorded diagnoses before pregnancy (epilepsy, pain, GAD)



Pregabalin and neurodevelopment study in the Nordics
Postnatal outcomes
- Attention deficit/hyperactivity disorder (ADHD)
- Autism spectrum disorder (ASD)
- Intellectual disability

Pregabalin and neurodevelopment study in the Nordics
Statistical analyses
- Crude and adjusted hazard ratios (HRs)
- Robust cluster-adjusted 95% CIs
- Cox proportional-hazards regression adjusted using propensity score (PS) fine stratification
- Distinct PS model for each country and contrast
- Trimming of the non-overlapping areas of PS distribution based on the PS distribution among exposed
- Pooling of country-specific relative risk estimates in a meta-analysis with fixed effects for different countries
- Post-hoc analysis using Mantel-Haenszel (MH) pooling (retaining zero-exposed)
Pregabalin and neurodevelopment study in the Nordics
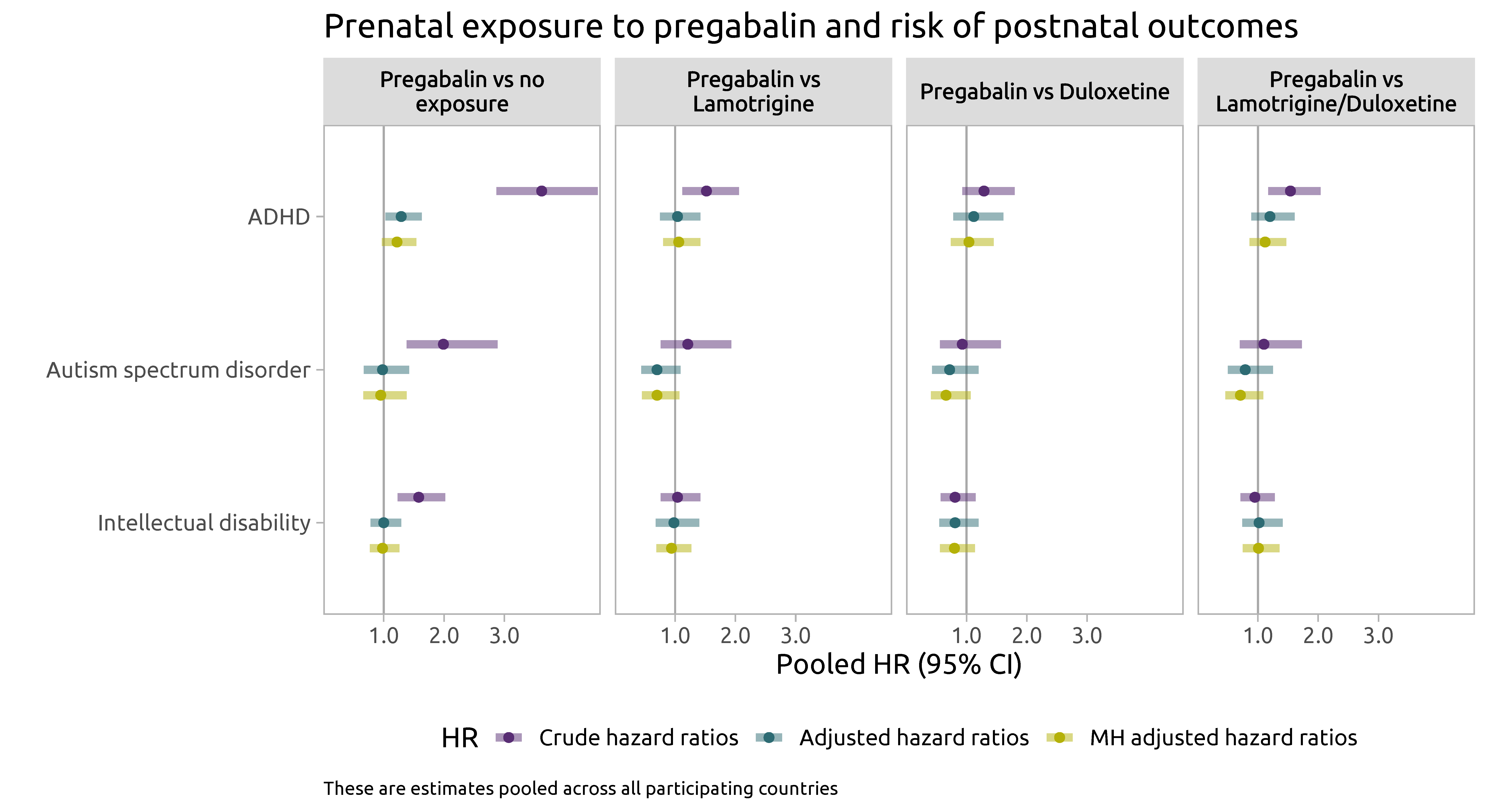
Main results
Pregabalin and neurodevelopment study in the Nordics
Additional results
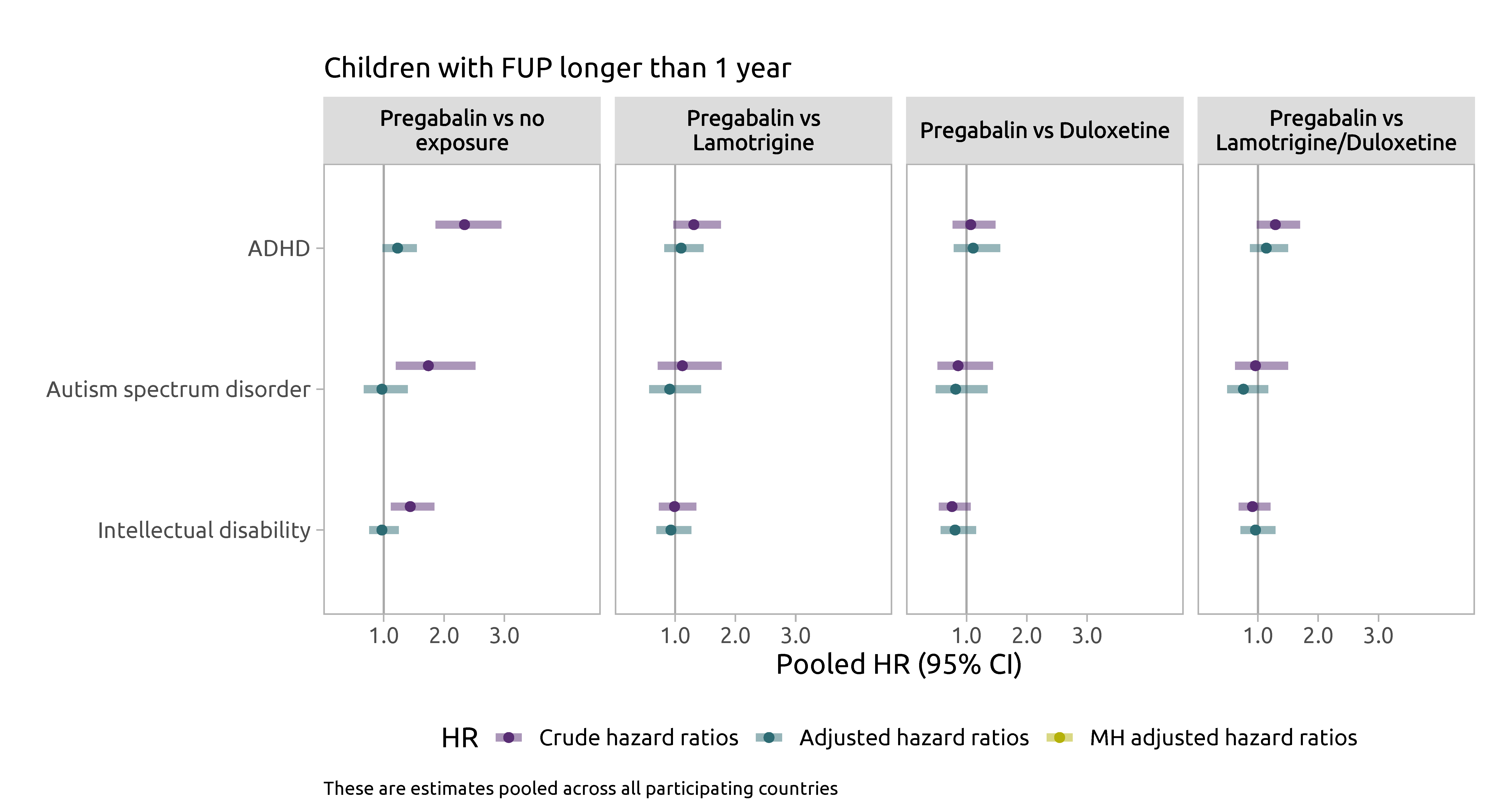
Pregabalin and neurodevelopment study in the Nordics
Monotherapy analyses
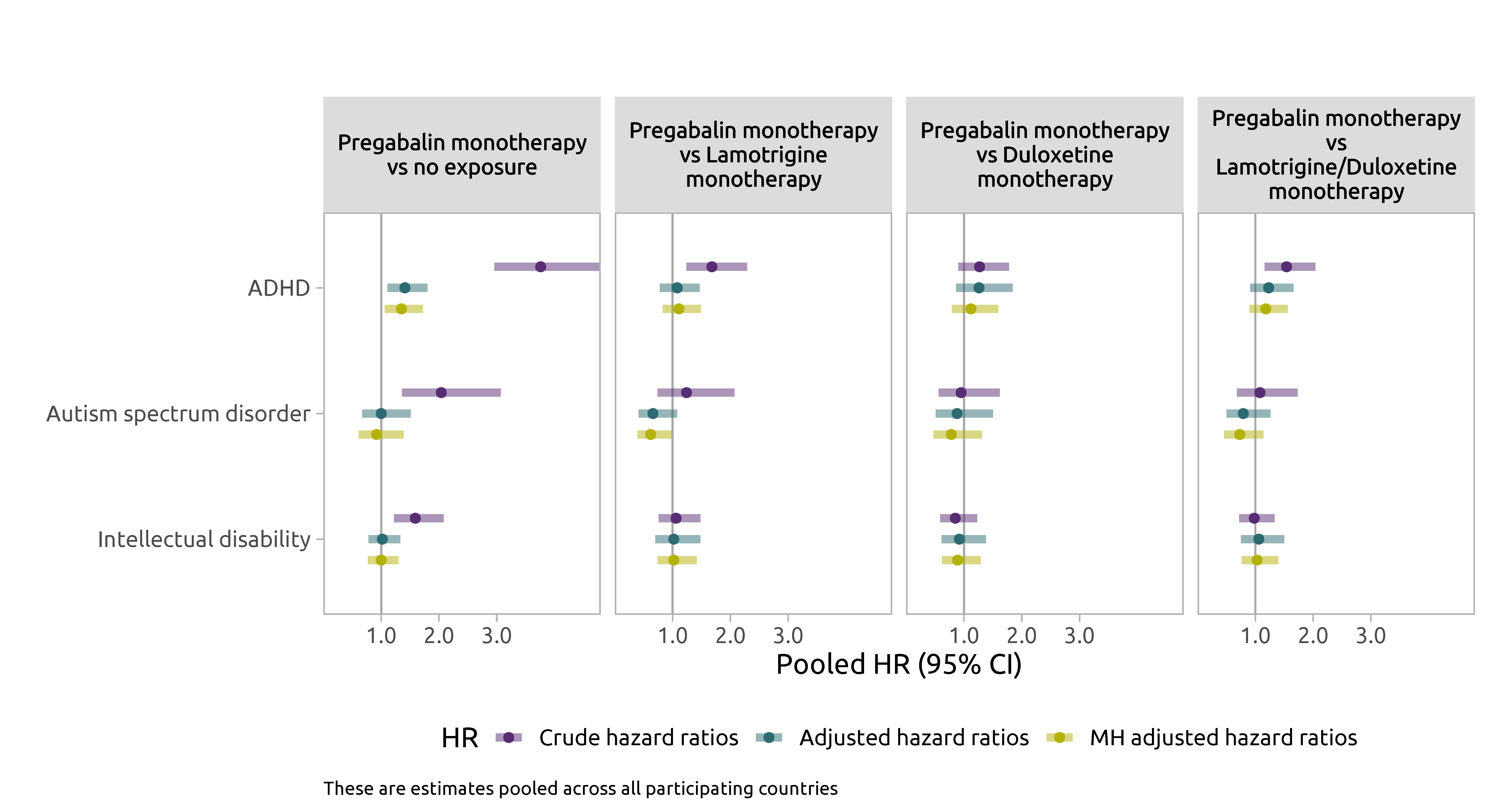
Results summary
- There was a 1.2 to 1.3-fold increased risk of SGA and ADHD among offspring with prenatal exposure to pregabalin vs no exposure to AED
- Associations attenuated in analyses using active comparators
- At least partly can be explained by residual confounding
- No evidence of an association between prenatal exposure to pregabalin and increased risk of ASD or intellectual disability
Pregabalin and neurodevelopment study in the Nordics
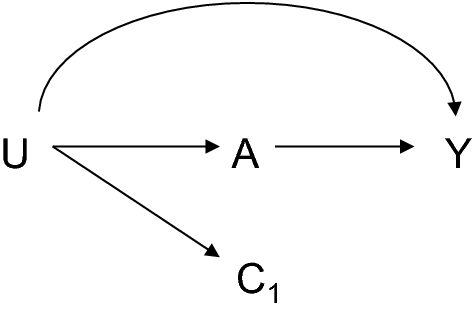
Residual confounding
- Unbalance for the contrast of offspring with exposure to pregabalin vs no exposure
![]()
- analgesics
- antipsychotics
- antidepressants
- history of depression and other neurological disorders
- Residual confounding by indication cannot be ruled out
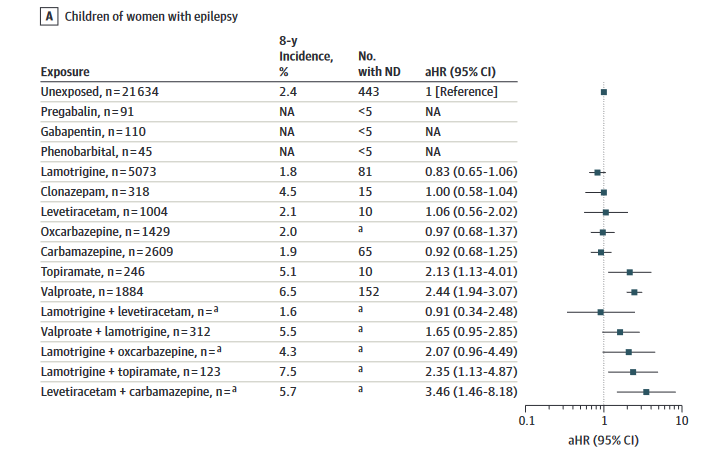
Pregabalin and neurodevelopment study in the Nordics
Comparisons with previous studies
- Valproate is associated with 1.5-fold increased risk of ADHD and 2.4-fold increased risk of any postnatal neurodevelopmental disorder1
Pregabalin and neurodevelopment study in the Nordics
Outcome misclassification
- Few false-positives are expected
- Positive predictive values for ADHD coding 87% (DK)
- Positive predictive values for autism coding 94% (DK)
- Age at ADHD onset:
- 8 years in Denmark
- 7.5 years in Finland
- 10.5 years in Norway
- 12 years in Sweden
- Children with subdiagnostic symptoms
Pregabalin and neurodevelopment study in the Nordics
Exposure misclassification
- Possible: true intake of pregabalin and comparators is unknown
- Redeemed prescriptions > issued prescriptions

Conclusion
- Increased risks in excess of 1.8 were unlikely for ADHD
- There was no evidence of an association with other examined neurodevelopmental postnatal outcomes
- Overall more reassuring finding than not